Notebooks#
Note
In this repository, all notebooks have been converted to MyST Markdown files
(with a .md extension) since this format is easier to visualize on GitHub.
This also makes it easier to view differences between versions of the notebooks
on the GitHub interface. To open these files as Jupyter Notebooks, you need to
have the Jupytext package installed (this will be installed automatically if you
run python -m pip install -r requirements.txt or
conda create -f environment.yml from this repository as outlined in Python setup & napari installation.)
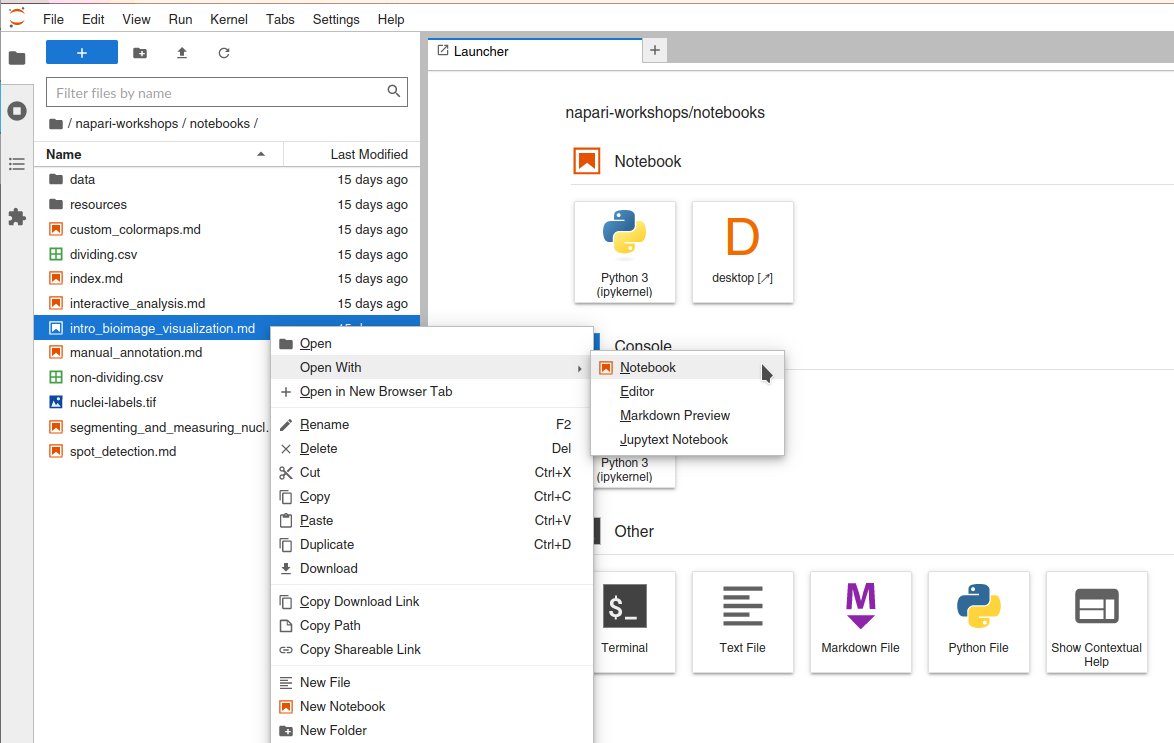
In the Jupyter notebook interface, if you right-click any of the .md files in
this folder now, you should see an option that says “Open with -> Notebook”
Alternatively, you can also convert the .md files into .ipynb files by running
jupytext –to ipynb <notebook_file>.md
in the command line.
Contents#
- Bioimage visualization in Python
- Interactive analysis with Jupyter notebook, napari, scikit-image, and scipy
- Segmenting nuclei with the
stardistnapari plugin - Bonus: Quantify nuclei shape
- Mitonet segmentation
- Manual annotation
- DIY Interactive Segmentation with napari
- Using custom colormaps in napari
- Conclusions
- Introduction to animation with napari