Basic image processing#
!pip install git+https://github.com/guiwitz/napari-skimage.git
!pip install napari-skimage-regionprops
import napari
from napari.utils import nbscreenshot
from napari_skimage.skimage_filter_widget import median_filter_widget
from napari_skimage.skimage_threshold_widget import threshold_widget
from skimage import data
# Load a sample 3D grayscale image (like cells)
viewer = napari.Viewer()
sample_image = data.cells3d()[30, 1] # This will load a sample grayscale image from skimage
image_layer = viewer.add_image(sample_image, name='sample_cells')
# Create the widgets without parameters
laplace_widget = median_filter_widget()
threshold_widget_instance = threshold_widget()
# Add the widgets to the viewer's dock
viewer.window.add_dock_widget(laplace_widget, area='right')
viewer.window.add_dock_widget(threshold_widget_instance, area='right')
# Capture the screenshot for the notebook
screenshot = nbscreenshot(viewer)
screenshot
Downloading file 'data/cells3d.tif' from 'https://gitlab.com/scikit-image/data/-/raw/2cdc5ce89b334d28f06a58c9f0ca21aa6992a5ba/cells3d.tif' to '/home/runner/.cache/scikit-image/0.24.0'.
What other tools does napari-skimage expose?#
filters
thresholding
binary morphology (erosion, dilation, opening, closing)
image math
rolling ball operations
find maxima
marching cubes
Activity#
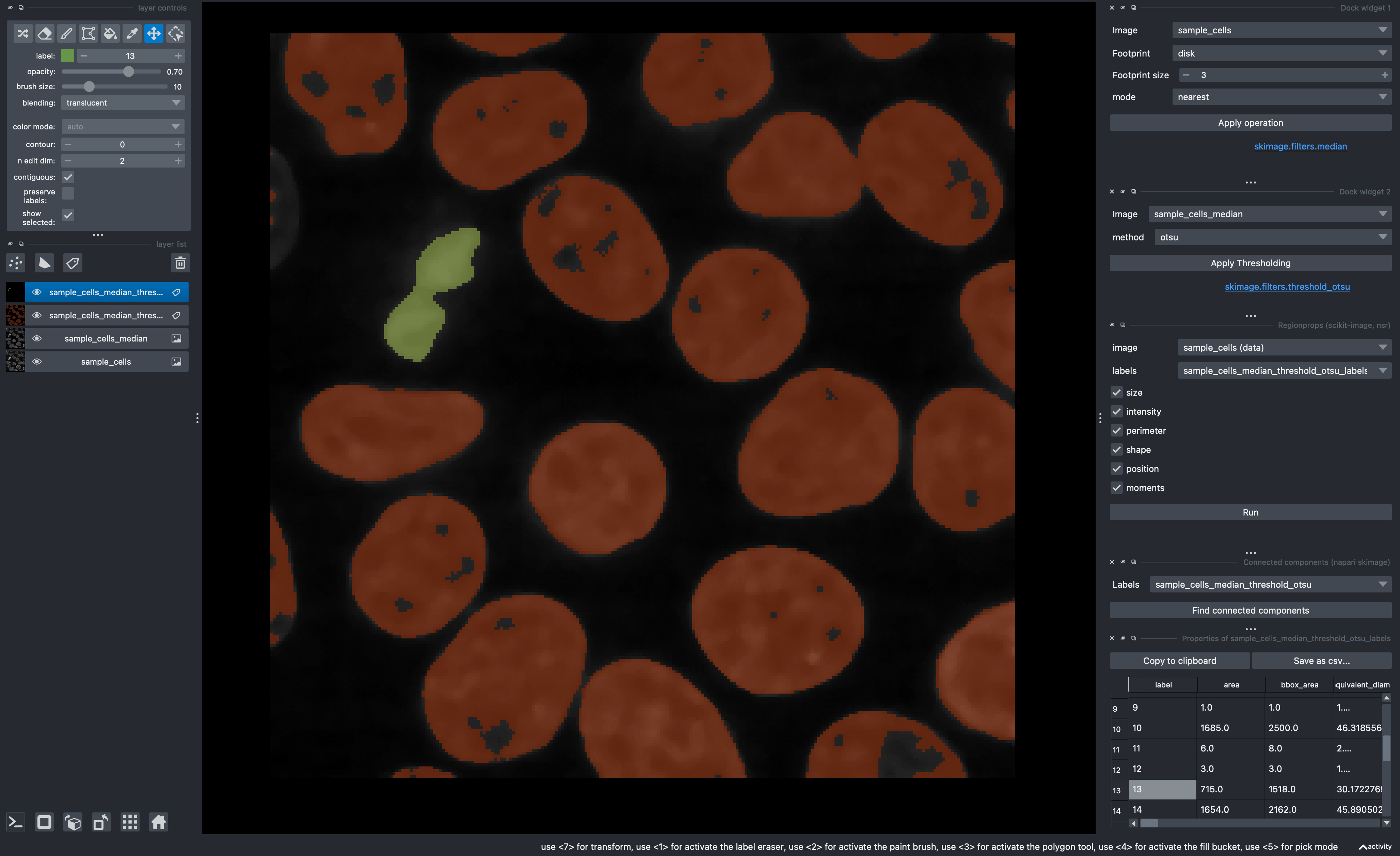
Use napari-skimage to do a vanilla segmentation of the cells image and display their statistics:
Use your favorite filter (median filter maybe?)
Use your favorite thresholding method (perhaps Otsu?)
Get the connected components
Use napari-skimage-regionprops (Note that this is under the
Toolsmenu asMeasurement Tables>Regionprops)Turn on the
Show selected checkboxfrom the Labels layer controls